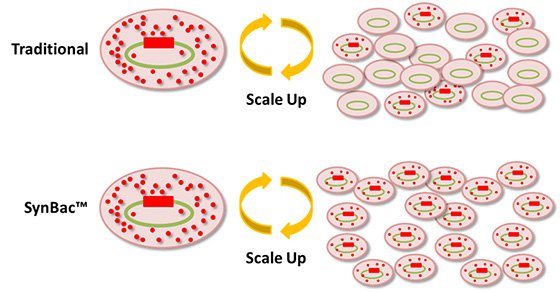
Traditional process scale up using baculovirus is notoriously problematic, with yields dropping progressively during scale up. SynBac™ are minimized baculoviral genomes created using modern synthetic biology techniques. The SynBac project aims to transform fermenter-scale baculoviral production of drug targets, and manufacturing scale production of biopharmaceuticals by creating minimal baculoviral genomes displaying exceptional DNA stability. In addition to improved DNA stability, our minimized genomes form the starting point for applicaions that require large DNA cargo delivery to mammalian cells, primary cells, or organs.
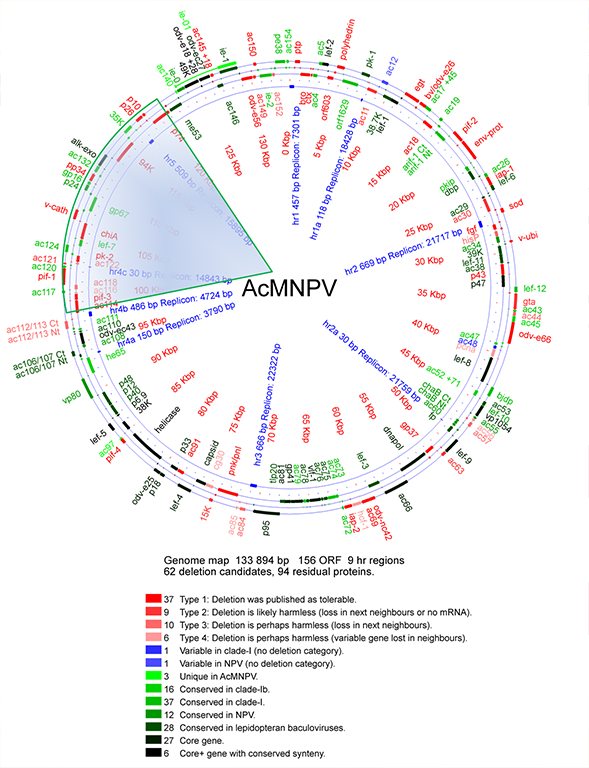
EMBL and IStLS carried out a comparative genome analysis and intense data mining to identify regions of the baculovirus genome that are potentially harmful for DNA stability, or are otherwise non-essential and can be discarded or re-engineering (see circle map below). Large sections of the AcMNPV genome are presently being rewired and tested for critical parameters such as expression yield and DNA stability. In an iterative process, improved genomes are being used as templates for further rounds of genome minimization and optimization.
Click here to download SynBac Manual v2.0
Use MultiBac™ protocols (click here) to assemble SynBac™ Viruses
Click here for DNA vector sequences
Click here to download Cre-ACEMBLER User Manual 3.0
Click here to download Cre-ACEMBLER Short Description
Click here to download Cre-ACEMBLER for Windows
Click here to download Cre-ACEMBLER for Linux
Click here to download SynBac reference 1
Click here to download baculovirus DNA instability reference 1
Click here to download baculovirus DNA instability reference 2
Click here to download baculovirus DNA instability reference 3
Below is a schematic representation of the AcMNPV genome (circle map). To provide an example, the rewiring target of the SynBac1.0 genome is indicated by the shaded triangle.